Tomato Epigenome Database |
Introduction to Illumina SequencingThe most crucial weapon in our arsenal to exploit the rich genetic and epigenetic resources of the Solanacea family is the Illumina sequencing machines. They are second-generation sequencing machines using the DNA sequencing by synthesis technology and could faithfully deliver gigabytes of sequencing data per week.

Sequencing library constructionTo sequences a sample of interest, we have to convert the input material into a "library" readable for the sequencing machine. This "library" is actually a mixture of short DNA fragments with specific linker sequences at both ends. If it is a genomic DNA sample, we will first break the long genetic DNA into 200 bp short fragments using Bioruptor or Covaris S2. In the case of RNA samples, they would also have to be fragmented and converted to short double-stranded DNA fragments. Only the chromatin-immunoprecipitation (ChIP) samples do not require further manipulation since they have already been fragmented before immunoprecipitation. These short DNA fragments also need to be end-polished using T4 DNA polymerase and polynucleotide kinase, which leave blunt-end with 5' phosphate. Then an "A" overhang is added to the 3' end using Klenow exo-, which force them to ligate to the Y-shape adapters with "T" overhang rather than joining each other.
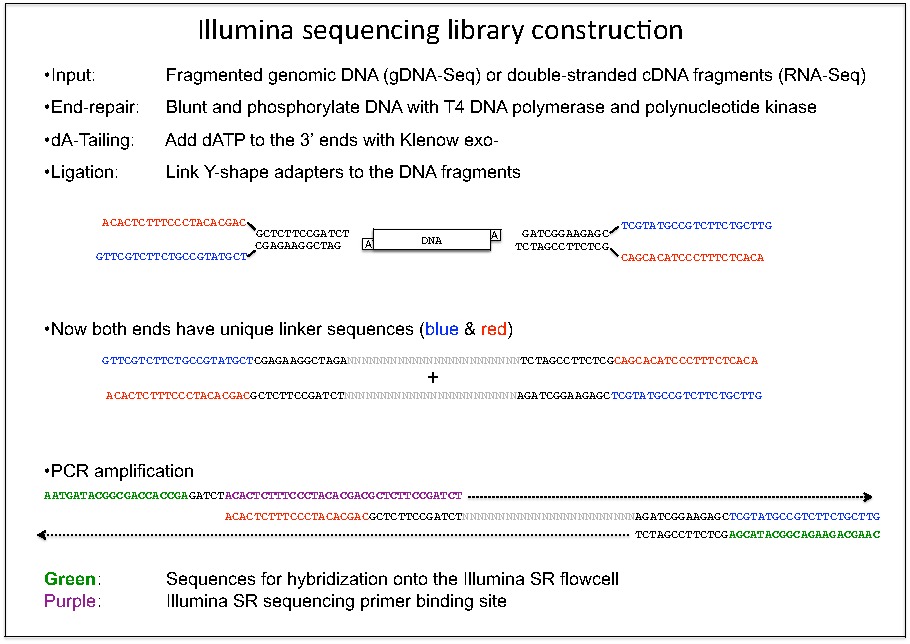
The ligation and the subsequent size fractionation is the most critical step of library construction. The Illumina GAIIx machine works best with tiny clusters of identical size, while HiSeq2000 tend to tolerate various library size. If we load a non-uniform library with a broad range of fragment size, it is most likely that the pass filter cluster count will drop below the 30 million per lane mark on GAIIx. Hence, for the low throughput DNA-Seq, bisulfite-Seq and RNA-Seq libraries, we would run the library into agarose gel using very high voltage (300 V) with lithium borate buffer (FBM LB buffer). We then stained the gel using SYBR-safe dye and carefully cut the gel slice containing the library. Ultraviolet (UV) light could damage the DNA sample, so Axel and Yun-ru have built some blue-light using LED with emission at 450 nM to excite the SYBR-safe dyes.

However, for our transcriptome sequencing projects, it is not possible to use such a low-throughput method (agarose gel) to prepare hundreds or even thousands of strand specific RNA-Seq libraries. For those, we use carboxyl group coated magnetic particles to purify and subsequently size select the libraries on 96-well plates. This method enables us to generate strand-specific transcriptome profiles for rice, tomato, cassava and sorghum at single nucleotide resolution.
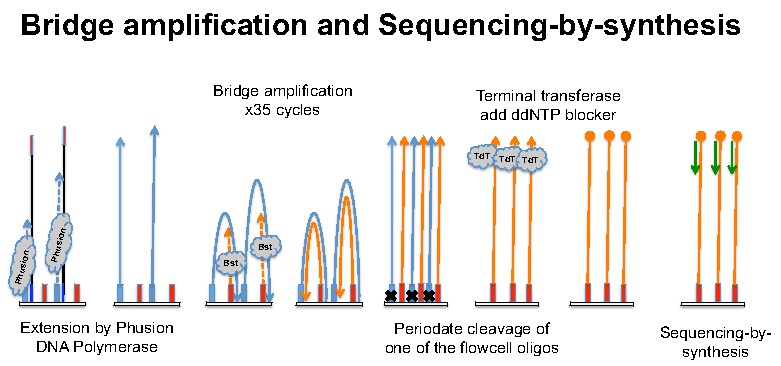
Cluster generation and sequencing by synthesisTo perform bridge-amplification, the denatured and diluted libraries are loaded to the flowcell, where they hybridize with the two surface oligos. The oligos on the paired-end (PE) and single-read (SR) flowcells are slightly different. PE and SR (including small RNA-Seq) libraries are constructed with different adapters and PCR primers. Figure 5 shows that PE libraries are compatible with SR flowcell but the adapters on SR and small RNA libraries can't hybridize to the PE flowcell.

On the cluster station or C-Bot, the original libraries (single stranded DNA) will be copied onto the SR flowcell by Physion DNA polymerase (some old version cluster kits use Taq) using one of the flowcell oligo as primer. The old template and Phusion will be washed away and the newly synthesized DNA contains adapter sequence complementary to the second flowcell oligo at its 3' end. So it can server as a template for primer extension by Bst DNA polymerase (bridge amplification). The whole idea of bridge amplification is to generate lots of copies of the original library immobilized on the flowcell surface. These identical DNA fragments (still have their complementary strand nearby) will form "clusters" and after one strand removed by periodate cleavage of the diol containing flowcell oligo. Now, the flowcell with millions of clusters can be moved to the GAIIx machine and sequenced by synthesis.
|
Fei Bioinformatics Lab, Boyce Thompson Institute and USDA Robert W. Holley Center |