Strand-specific RNA Sequencing
Cold Spring Harb Protoc; 2011; doi:10.1101/pdb.prot5652
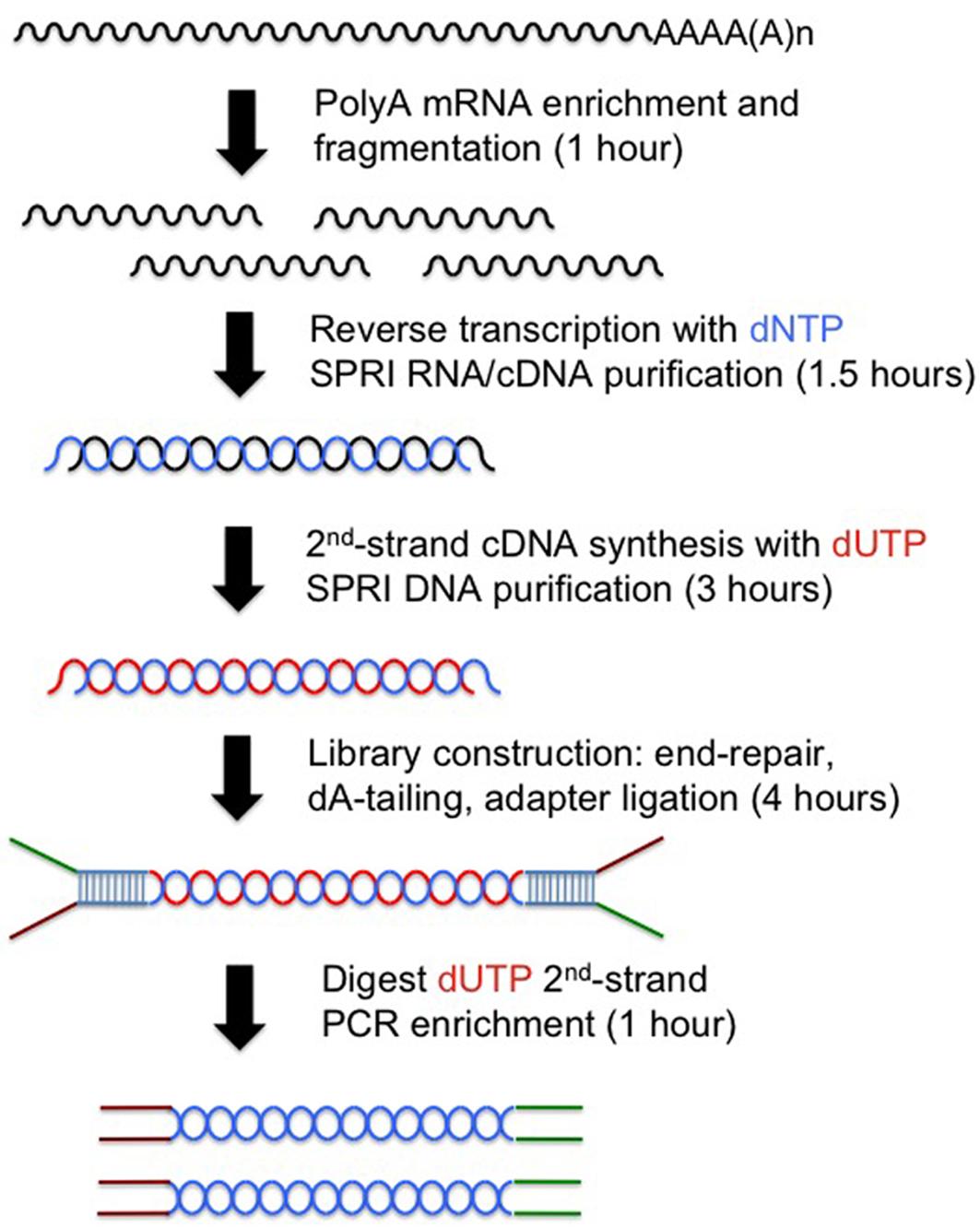
PolyA RNA isolation and Fragmentation (1 hour)
- 1. Prepare appropriate amount of oligodT25 Dynabeads (20 uL per sample) in 1.5 mL tube, wash twice with 200 uL of 1x Binding Buffer.
- 2. Resuspend Dynabeads in appropriate volume of 2x Binding Buffer (50 uL per sample), pipette 50 uL of beads to each 0.2 mL PCR tube.
- 3. Add 2-5 ug total RNA (in 50 ul H2O, so it contains 1x binding buffer), mix well.
- 4. Close the tubes, heat to 65C for 1 min on a thermocycler with heated lid and incubate at RT for 5-10 min with occasional shaking.
- 5. Briefly spin the PCR strip to collect the beads on the cap, place on magnetic stand, remove the solution, wash twice with 150 uL of Washing Buffer.
- 6. Elute the mRNA by adding 50 uL TE and incubate at 70 C for 1 min. immediately put the strip back on ice.
- 7. Transfer the 50 uL elute (TE) to a new strip with 50 uL 2 x binding buffer (so the final concentration is 1x binding buffer).
- 8. Wash the used beads with 100 uL of H2O, resuspend in 20 uL 1x binding buffer and transfer the 20 uL beads to the tube with RNA. Now repeat the binding and washing step 4 and 5.
- 9. Wash again with 30 uL 1x pre-chilled SuperscripIII buffer to prevent carries over of salt and detergent. Note: lithium inhibits reverse-transcriptase.
- 10. Resuspend the beads in 10 uL of superscript buffer:
| SuperscriptIII first-strand buffer | 4 ul |
| Random Primer (1 ug/ul) | 1 ul |
| Oligo dT VN (100 ng/uL) | 1 ul (optional for 3’RNA seq) |
| 0.1 M DTT | 1 ul |
| H2O | to 10 ul |
- 11. Incubate at 94C for exactly 5 min to fragment the mRNA, immediately place on ice.
- 12. Snap spin the strip, place on magnetic stand, and transfer the solution containing fragmented mRNA to a new strip. Note: stopping point, eluted RNA can be stored at -80 C.
First-strand cDNA synthesis
- 13. Assemble the following RT reaction.
| Fragmented mRNA in 2x RT buffer | 10 uL |
| RNasin Plus | 0.5 uL |
- 14. Incubate at 25 C add the following RT master mix.
| H2O | 6.88 u |
| Actinomycin D (1 ug/ul) | 0.12 uL |
| DTT (100 mM) | 1 uL |
| dNTP (10 mM) | 1 uL |
| Superscript III | 0.5 uL |
- 15. Perform RT reaction as:
- 16. Immediately 36 uL RNAClean XP to each tube and incubate the mixture on ice for 15 min. Note: the solution is very viscous; pipette up and down at least 10 times to mix.
- 17. Collect the SPRI-beads on magnetic stand. Note: when use SPRI-beads, always keep the PCR strip/plate on the magnet till the final elution step.
- 18. Wash twice with 75% EtOH without disturbing the beads.
- 19. Air-dry the beads for 2 min and elute RNA/cDNA hybrid with 10 uL H2O.
Second-strand synthesis with dUTP.
- 20. Prepare the 2nd strand reaction mastermix on ice as follow:
| 10x Blue Buffer (or NEB buffer 2) | 1.5 uL |
| 10 mM dNTP mix (dA, dC, dG and dUTP) | 1 uL |
| RNase H (5 U/uL) | 0.2 uL |
| DNA polymerase I (10 U/uL) | 1 uL |
| H2O | 1.3 uL |
- 21. Add 5 ul of the mastermix to each 10 ul RNA/cDNA, incubate at 16C for 2.5 hours. Note: the completed 2nd strand reaction can be held in the PCR machine at 4C overnight.
- 22. Purify dsDNA using 1.8 volumes of AMPure XP beads and elute with 10 uL of H2O. Note: stopping point, eluted dsDNA can be stored at -20 C.
End-repair
- 23. Prepare appropriate amount of the end-repair mastermix on ice as follow:
| 10x end-repair buffer (or NEB PNK buffer) | 1.5 uL |
| 10 mM dNTP mix | 0.5 uL |
| H2O | 2.5 uL |
| End Repair Enzyme mix LC | 0.5 uL |
- 24. Add 5 uL of the mastermix to 10 uL of the dsDNA, incubate at 20C for 30 min.
- 25. Purify using 1.8 volumes of AMPure XP beads and elute with 10 uL of H2O.
dA-tailing
- 26. Prepare appropriate amount of the mastermix on ice as follow:
| 10 x Blue Buffer (or NEB buffer 2) | 1.5 uL |
| 10 mM dATP | 0.5 uL |
| H2O | 2.5 uL |
| Klenow 3'-5' exo | 0.5 uL |
- 27. Add 5 ul of the mastermix to each sample and incubate at 37C for 30 min.
- 28. Purify dsDNA using 1.8 volumes of AMPure XP beads, elute with 8 uL of H2O.
Y-shape adapter Ligation
- 29. Add 0.5 uL of the desired barcode adapter (15 uM) to each sample.
- 30. Prepare the mastermix on ice as follow:
| 2x Rapid ligation buffer | 8.5 uL |
| T4 DNA Ligase HC (600 U/uL) | 0.5 uL |
- 31. Add 9 uL of the mastermix to each well, mixed by pipetting up and down and incubate at RT for 15 min. Note: stopping point, the ligated library can be stored at -20 C.
Triple-SPRI purification and size selection
- 32. Add 1 volume (18 uL) of AMPure XP beads, incubate 10 min at RT to purify the ligation product, elute with 10 uL TE.
- 33. Add 1.4 volume (14 uL) of AMPure XP beads, incubate 10 min at RT to purify the ligation product, elute with 10 uL TE.
- 34. To perform size selection, add exactly 0.8 volume of AMPure XP beads (8 uL) to the eluted DNA and incubate at RT for 5 min.
- 35. Pull the beads to the side of the plate/tube on a magnetic-stand and carefully pipette the supernatant (18 uL) to a tube with 10 uL of AMPure XP beads.
- 36. Mixed by pipetting, incubate 10 min at RT.
- 37. Collect the beads on magnetic stand and wash twice with 75% ethanol without disturbing the beads.
- 38. Elute the DNA with 10 uL water. Note: stopping point, the size-fractionated DNA can be stored at -20 C.
PCR enrichment
- 39. Digest the 2nd strand DNA with 0.5 uL of Uracil DNA Glycosylase at 37 C for 15 min.
- 40. Prepare PCR reaction as follow:
| UDG digested DNA | 5 uL |
| Primer mix (10 uM each) | 1 uL |
| 5x Phusion HF Buffer | 5.5 uL |
| 10 mM dNTP | 0.5 uL |
| H2O | 7.5 uL |
| Phusion II | 0.5 uL |
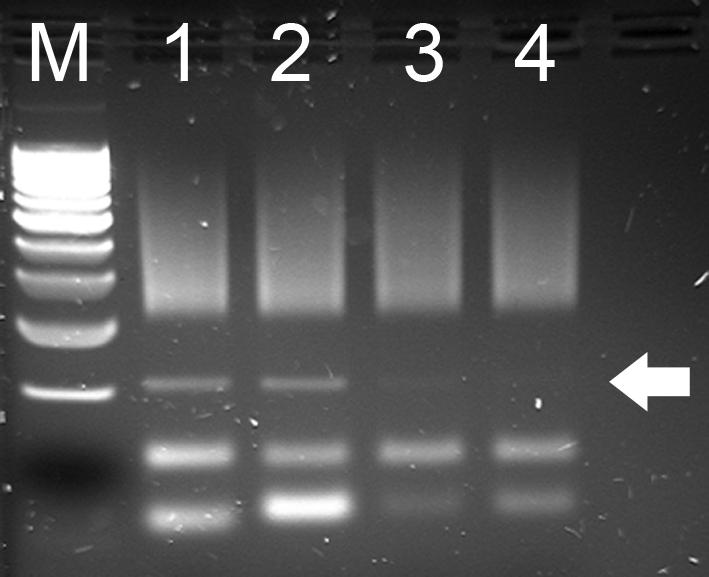
- 41. Randomly select a few libraries and run 2 uL of their pre-purified PCR product on agarose gel to check whether there is adapter dimer contamination (around 100 bp, indicated by arrowed).
- 42. Purify using 1.2 volumes of AMPure XP beads, elute with 20 uL of TE.
Mix barcoded libraries for multiplexed sequencing
- 43. Measure the DNA concentration of each library using Quan-IT DNA HS assay kit (single-tube).
- 44. Combine equal amount (e.g. 20 ng) of each barcode library.
- 45. Concentrate the library using 1.2 volume of AMPure XP and elute with 10 uL TE.
General procedure for using AMPure beads
- 1. Add appropriate amount of beads to the sample as indicated in the protocol.
- 2. Mix well by pipette up and down for 10 times.
- 3. Incubate the plate/tube at RT for 15 min. Note: unless specified in the protocol, prolong or low temperature incubation will result in binding of small DNA fragments such as adapter dimers.
- 4. Place on the magnet stand for 2-5 min to collect the beads. Note: viscous solution will require longer collection time.
- 5. Gently remove the solution without disturbing the bead pellets.
- 6. Add 200 uL of 75% ethanol. Note: keep the plate on the magnet.
- 7. Wait for 30 s and remove the ethanol without disturbing the bead.
- 8. Repeat the 75% ethanol wash once more. Note: inspect the plate carefully and remove any remaining ethanol droplets.
- 9. Air-dry for 1-2 min and add appropriate amount of water (or TE) to elute the DNA. Note: do not over dry the beads.
- 10. Place on the magnet, transfer elute to a new plate/tube.
|
